COVID-19 Testing R&D Highlights

Developed analysis tools to maintain awareness of the evolution of the SARS-CoV-2 RNA genome, as it relates to nucleic acid-based assays. For example, see https://covid19.edgebioinformatics.org/#/home for an automatic in silico evaluation of diagnostic assays used around the world. On December 20, 2020, this web application provided comparisons for 17 assays using 44,224 SARS-CoV-2 genomes.
Identified distinguishing signatures in the SARS-CoV-2 RNA genome that can be used to rapidly detect this pathogen and other co-infecting pathogens in multiplexed assays. For example, this advance identified potentially significant relationships of co-infections with Streptococcus pneumonia and Streptococcus pyogenes in positive COVID-19 samples; co-infections with other pathogens are being evaluated.
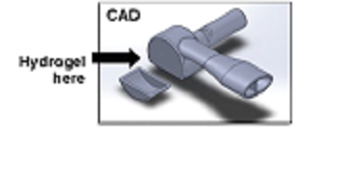
Developed a small nucleic acid test instrument that can rapidly detect SARS-CoV-2 with high sensitivity. The Reveal-CoV instrument, which uses a one-step amplification process followed by heat inactivation and lysis, employs a colorimetric change for detection.

Devised a “COVID Whistle” that incorporates sample collection media to simplify both collection of exhaled breath and extraction of virus; device is being evaluated by a university collaborator.